Research highlights July-Sept 2018
Recent Posts
Spotlight on: Melanie Eckersley-Maslin 25 November 2022
Spotlight on: Jessamy Tiffen 25 July 2022
ECR Spotlight on: Alex Woodworth 16 July 2022
Epigenetics 2022: Four weeks left for poster abstract submissions! 15 July 2022
Emma Whitelaw ECR Publication Award 30 June 2022
Epigenetics 2022 – Meet the speakers 17 June 2022
ECR Spotlight on: Kate Giles 31 May 2022
Spotlight on: Luciano Martelotto 30 May 2022
Epigenetics 2022 speakers announced 20 April 2022
Spotlight on: Hamish King 18 April 2022
ECR Spotlight on: William Schierding 6 April 2022
Spotlight on: Louise Bicknell 14 March 2022
Categories
News (57)
Opportunities (3)
Publications (7)
Spotlight on … (16)
World epi news (2)
Archived news
2022 (12)
2021 (9)
2020 (9)
2019 (1)
2018 (6)
2017 (6)
2016 (6)
2015 (17)
2014 (1)
by AEpiA | Oct 30, 2018 | News
Here are the latest highlights of Australian epigenetics publications between July-Sept 2018:
Inhibition of a K9/K36 demethylase by an H3.3 point mutation found in paediatric glioblastoma (Published: 07 August 2018) This study, led by Associate Professor Lee Wong at Monash University in Melbourne, reveals that oncogenic histone point mutations can “exert their effect through interaction with a range of epigenetic readers, writers and erasers”. Looking at the G34R (glycine to arginine) substitution mutation in the histone variant H3.3, the authors report downstream widespread changes in H3K9me3 and H3K36me3. The H3.3 G34R mutation is found in paediatric gliomas and the authors unravel that the changes to H3K9me3 and H3K36me3 are because H3.3 G34R is able to bind the demethylase KDM4 and inhibit its enzymatic activity. Inhibition of KDM4 activity in this way then affects H3K9me3 and H3K36me3 activity. Indeed the H3K9me3 and H3K36me3 profile in H3.3 G34R mutants in mouse embryonic stem cells is shown to be similar to KDM4 A/B/C triple-knock down. Thus, the study identifies the KDM4 histone lysine demethylases as “the key chromatin modifiers, which are disrupted by this mutant histone” and demonstrates how histone point mutation in cancer may exert their effects by interacting with epigenetic readers, writers and erasers.
In this year’s September edition of Nature Structural and Molecular Biology, Associate Professor Marnie Blewitt’s laboratory revealed that the noncanonical SMC protein Smchd1 is a novel regulator of long-range chromatin interactions in mice. The study, conducted primarily at the Walter and Eliza Hall Institute of Medical Research in Melbourne, tested the role of Smchd1 in maintaining chromosome structure using in situ Hi-C in Smchd1 depleted female mice. In doing so the authors discovered a role for Smchd1 in regulating Hox clusters in particular -
thus adding to the canon of proteins known to epigenetically regulate Hox gene silencing during development. The report focused on the effect of losing Smchd1-dependent chromatin interaction in the inactive X (Xi) chromosome. This showed an increase in X-linked chromatin interaction in the absence of Smchd1, without the expected increase in reactivation of genes from the Xi. Instead, the report reveals, “spreading of trimethylated histone H3K27 domain into regions not normally decorated by this mark” on Smchd1 depleted Xi. The authors conclude that Smchd1 is able to insulate chromatin to limit access to other chromatin-modifying proteins and thereby regulate long-range chromatin interactions, thus adding to our understanding of how chromatin architecture is regulated. For the Full-Text Article: https://www.nature.com/articles/s41594-018-0111-z
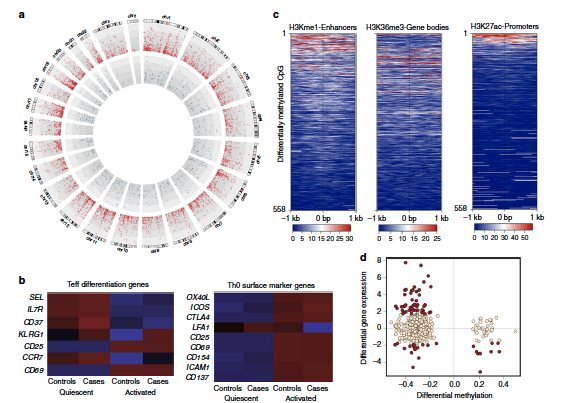
Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy (Published: 17 August 2018) From Melbourne’s Murdoch Children’s Research Institute, Professor Richard Saffery and colleagues have published an investigation on naïve CD4+ T cell activation in a cohort of infants with egg allergy. The study design involved activating T cells from non-atopic (control) and atopic individuals under non-differentiating conditions, and then profiling DNA methylation, gene expression and cell proliferation in these cells. The authors gained insight into naïve T cell responsiveness to activation. Comparison between the two groups showed that infants with food allergies have gene dysregulation in their T cell activation pathways leading to
poor lymphoproliferative responses. The study reveals that the suboptimal T cell responses are underpinned by “reduced expression of cell cycle–related targets of the E2F and MYC transcription factor networks, and remodeling of DNA methylation at metabolic (RPTOR, PIK3D, MAPK1, FOXO1) and inflammatory genes (ILIR,
ILI8RAP, CD82)”. The pathway modifications by gene-environment interactions seen in food allergy are thereby connected to epigenetic dysregulation in the early stages of signal transduction from the T cell receptor complex. For the Full-Text Article: https://www.nature.com/articles/s41467-018-05608-4
The state of long non-coding RNA biology (Published: 10 August 2018) This June, the journal non-coding RNA published a review by Professor John Mattick, who has recently moved from the Garvan Institute of Medical Research in Sydney to Genomics England in London. In the review titled ‘The State of Long Non-Coding RNA Biology,’ Professor Mattick states that long non-protein-coding RNAs (lncRNAs) “are still to be widely accepted as major players in gene regulation.” This, he asserts, is “despite well characterized examples, their scaling with developmental complexity, and many demonstrations of their association with cellular process, development and diseases.” The review shines the spotlight on lncRNAs, starting with the history of their discovery and continuing to go into detail of the structure-function relationships of IncRNAs, which the review claims is “the most pressing challenge” to resolving the IncRNAs functional repertoire. The review ends by making the statement that RNA is “not (simply) . . . a transient intermediate between gene and protein, but rather the central computational engine of cell biology, differentiation and development, brain function and perhaps even evolution itself” and predicting that “many textbooks may have to be rewritten once the full dimensions of regulatory RNA biology are revealed.” For the Full-Text Article: https://doi.org/10.1111/cea.13031
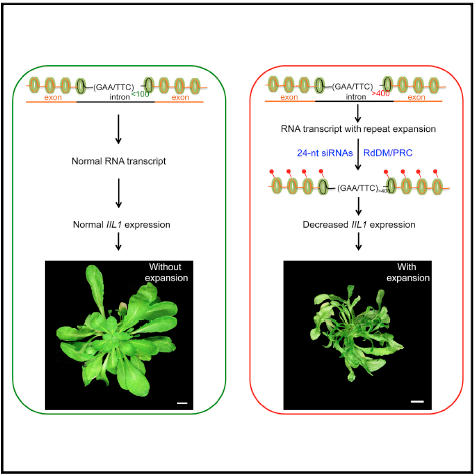
From Melbourne’s Monash University, this study delves into transcriptional downregulation caused by intronic triple expansions. Expansion of trinucleotide repeats in intronic regions, as seen in diseases such as Friedreich’s ataxia and fragile X, can be expansive, with the possibility of up to 2,000 repeats. How these repeats lead to reduction in certain protein expression in these diseases was the interrogated question. Led by Associate Professor Sureshkumar Balasubramanian and published in Cell in August this year, the report revealed that: “triple repeat expansions can lead to local siRNA biogenesis, which in turn down regulates transcription”. The study goes on to show that this deregulation is mediated by RNA-dependent DNA methylation (RdDM) epigenetic
modifications. Using an IIL1 repeat expansion in Arabidposis Thaliana, the authors unravel this molecular mechanism for the transcriptional downregulation in Arabidopsis. The report points to the need to assess RNA-dependent transcriptional gene silencing pathways to further understand the epigenetic attenuation of protein expression, particularly for disease conditions such as Friedreich’s ataxia. https://www.nature.com/articles/ijo2017228
– Do you have any research news you would like to share with AEpiA members? Please email us about any recent publications, awards or events!