Research highlights Jan-March 2018
Recent Posts
Spotlight on: Melanie Eckersley-Maslin 25 November 2022
Spotlight on: Jessamy Tiffen 25 July 2022
ECR Spotlight on: Alex Woodworth 16 July 2022
Epigenetics 2022: Four weeks left for poster abstract submissions! 15 July 2022
Emma Whitelaw ECR Publication Award 30 June 2022
Epigenetics 2022 – Meet the speakers 17 June 2022
ECR Spotlight on: Kate Giles 31 May 2022
Spotlight on: Luciano Martelotto 30 May 2022
Epigenetics 2022 speakers announced 20 April 2022
Spotlight on: Hamish King 18 April 2022
ECR Spotlight on: William Schierding 6 April 2022
Spotlight on: Louise Bicknell 14 March 2022
Categories
News (57)
Opportunities (3)
Publications (7)
Spotlight on … (16)
World epi news (2)
Archived news
2022 (12)
2021 (9)
2020 (9)
2019 (1)
2018 (6)
2017 (6)
2016 (6)
2015 (17)
2014 (1)
by Madhavi Maddugoda| May 20, 2018 | News, Publications
We thought we’d share some brief highlights of Australian epigenetics publications from the beginning of this year (2018):
The Tumor Suppressor Hic1 Maintains Chromosomal Stability Independent of Tp53 In a brief communication to Oncogene, this January, Professor Watkins from the Garvan Institute of Medical Research, Sydney and colleagues from Monash University, Melbourne used a conditional knockout mouse model to inactivate Hypermethylated–in-Cancer1 (Hic1) tumour suppressor gene. Using this model, Szczepny et al., show that the knockout mouse phenocopies a Brca1 deletion. The knockout resulted in cell cycle arrest, premature senescence, chromosomal instability and spontaneous transformation in vitro. The study reveals that Hic1, which is inactivated by epigenetic silencing in many cancers, “may contribute to malignant transformation through the acquisition of chromosomal instability”, when expression is lost in early tumour formation.
For full-text by Szczepny et al., see – doi 10.1038/s41388-017-0022-1
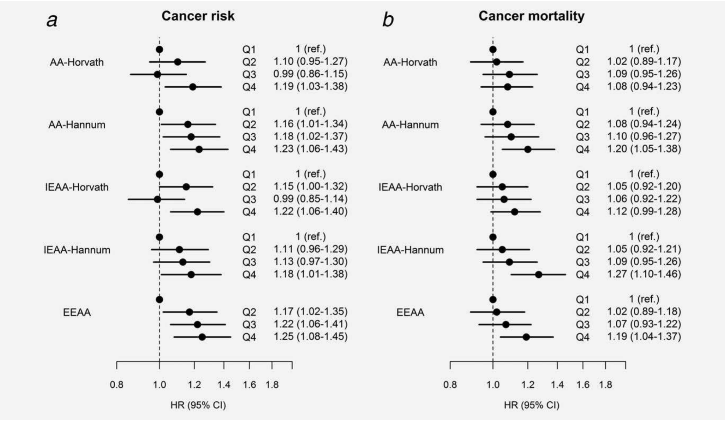
In this publication, Dr Pierre-Antoine Dugué et al., from Cancer Council Victoria, and The University of Melbourne utilize the Melbourne Collaborative Cohort Study to look for an association between ageing and cancer. For this, the authors compared ‘age acceleration’,
against the risk of and survival from colorectal, gastric, kidney, lung, prostate, urothelial cancer and B‐cell lymphoma. Age acceleration is a measure of biological ageing based on DNA methylation. Based on their analysis the study was able to identify that “DNA methylation-based measures of biological ageing are associated with increased cancer risk and shorter cancer survival, independently of major health risk factors”. Further, the study found that this was consistent across cancer types. As blood samples were obtained from participants prior to cancer diagnosis, the findings add evidence to the use of “methylation markers of biological ageing as putative predictors of health outcomes”. For full-text by Dugué et al., see – https://doi.org/10.1186/s13059-017-1302-3
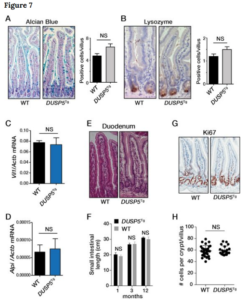
Based on their finding that the nuclear ERK-selective phosphatase DUSP5 is downregulated in colorectal tumours and cell lines Togel et al., investigate epigenetic changes to the DUSP5 promoter. The study finds that indeed a subset of DUSP5 promoters are methylated in colorectal cancer (CRC) cell lines and tumours. This promoter methylation is reported to be present particularly in CRCs with the CpG island methylator phenotype (CIMP), which is used for diagnostic,
Epigenome-Wide Analysis in Newborn Blood Spots from Monozygotic Twins Discordant for Cerebral Palsy Reveals Consistent Regional Differences in DNA Methylation
used Illumina Infinium Human Methylation 450 BeadChip arrays on DNA from 15 newborn monozygotic twin pairs. These twins later became discordant for Cerebral palsy. Their results revealed 33 differentially methylated probes related 2 Differentially Methylated Regions (DMRs) making “an initial step towards investigating potential CP-associated epigenetic differences, with the longer term aim of identifying predictive biomarkers with clinical utility.” For the full-text article by Mohandas et al., DOI: 10.1186/s13148-018-0457-4
prognostic and monitoring purposes. The study by Professor Mariadason and colleagues from the Olivia Newton-John Cancer Research Institute, and Ludwig Institute for Cancer Research in Melbourne concludes that this epigenetic change “could not account for reduced DUSP5 expression”, but it “can serve as an additional means of identifying CIMP-high colorectal cancer”. For the full-text article by Tögel et al., see – DOI: 0.1038/s41598-018-20176-9
From the Murdoch Children’s Research Institute, Melbourne and the Department of Paediatrics at The University of Melbourne, Mohandas et al., utilise the discordant monozygotic twin model to understand and measure epigenetic changes associated with the development of Cerebral palsy. The study led by A/Professor Craig
– Do you have any research news you would like to share with AEpiA members? Please email us about any recent publications, awards or events!