Australian epigenetics research news Oct – Dec 2017
Recent Posts
Spotlight on: Melanie Eckersley-Maslin 25 November 2022
Spotlight on: Jessamy Tiffen 25 July 2022
ECR Spotlight on: Alex Woodworth 16 July 2022
Epigenetics 2022: Four weeks left for poster abstract submissions! 15 July 2022
Emma Whitelaw ECR Publication Award 30 June 2022
Epigenetics 2022 – Meet the speakers 17 June 2022
ECR Spotlight on: Kate Giles 31 May 2022
Spotlight on: Luciano Martelotto 30 May 2022
Epigenetics 2022 speakers announced 20 April 2022
Spotlight on: Hamish King 18 April 2022
ECR Spotlight on: William Schierding 6 April 2022
Spotlight on: Louise Bicknell 14 March 2022
Categories
News (57)
Opportunities (3)
Publications (7)
Spotlight on … (16)
World epi news (2)
Archived news
2022 (12)
2021 (9)
2020 (9)
2019 (1)
2018 (6)
2017 (6)
2016 (6)
2015 (17)
2014 (1)
by Madhavi Maddugoda| Jan 22, 2018 | News, Publications
Here are some brief highlights of Australian epigenetics publications from the end of last year (2017):
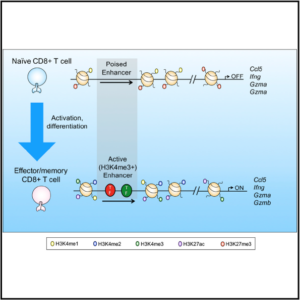
Regulation of H3K4me3 at Transcriptional Enhancers Characterizes Acquisition of Virus-Specific CD8+ T Cell-Lineage-Specific Function
– Do you have any research news you would like to share with AEpiA members? Please email us about any recent publications, awards or events!
In this study, Professor Turner and colleagues from Monash University and the Doherty Institute at the University of Melbourne have mapped the dynamic regulation of transcriptional enhancers (TEs) in T cells responding to an acute influenza A infection. By doing so they have identified key epigenetic mechanisms that underpin infection specific T cell differentiation, an essential
requirement for pathogen clearance. In their publication in Cell Reports the authors used ChIP-seq to map the histone dynamics of 25,000 putative CD8+ T cell transcriptional enhancers differentially utilized during virus-specific T cell differentiation. The study revealed the acquisition of a non-canonical (H3K4me3+) chromatin signature on a subset of dynamically regulated transcriptional enhancers unique to virus-specific CD8+ T cell differentiation. This identified “the genomic location for T cell lineage-specific transcription factor binding required for virus-specific T cell differentiation” and adds vital pieces to the puzzle of infection specific T cell regulation. For full text by Russ et al., see – http://www.cell.com/cell-reports/fulltext/S2211-1247(17)31779-5
The DNA Methylation Landscape of CD4þ T cells in Oligoarticular Juvenile Idiopathic Arthritis
From Murdoch Children’s Research Institute and the University of Melbourne, Professor Ellis and colleagues report “a lesser relevance of DNA methylation levels in childhood, compared to adult, rheumatic disease”. In their study, published in the Journal of Autoimmunity, the authors used Illumina HumanMethylation450 arrays to perform a genome-scale analysis of CD4. T cell DNA methylation of oligoarticular juvenile idiopathic arthritis patients and age and sex-matched controls. While adult autoimmune rheumatic diseases, such as rheumatoid arthritis, have been associated with altered DNA methylation, the article reveals that the pediatric autoimmune disease, oligoarticular juvenile idiopathic arthritis, does not show substantially altered methylation in oJIA in CD4. T cells. For full text by Chavez-Valencia et al., see – https://www.sciencedirect.com/science/article/pii/S0896841117305863
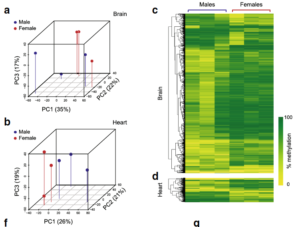
Isogenic Mice Exhibit Sexually-Dimorphic DNA Methylation Patterns Across Multiple Tissues In this study published in
published in BMC Genomics, A/Professor Suter and colleagues from Sydney’s Victor Chang Cardiac Research Institute aimed to understand the extent to which epigenetic states are influenced by sex; given sexual dimorphism is relevant to so many diseases. For this, the authors
used DNA methylation patterns from multiple tissues of isogenic male and female mice. Using this model the authors were able to identify “thousands of sexually dimorphic loci”, which they report to be “largely autonomous to each tissue”. The paper reveals that sex influences methylation patterns in a tissue-specific manner and therefore suggests that at least some of the phenotypes that carry gender bias are derived from gender differences in underlying epigenetic states. For full text by McCormick et al., see – https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-017-4350-x
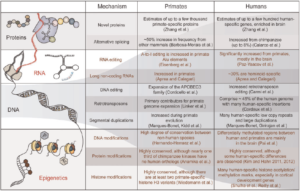
Multiple Innovations in Genetic and Epigenetic Mechanisms Cooperate to Underpin
Human Brain Evolution
In their Perspective, Dr Guy Barry and Mainá Bitar explore how the human brain differs from those of other species, with a focus on evolutionary adaptations and functionality. Given only 1% of the human genome is unique compared to a chimpanzee,the authors from Brisbane’s QIMR
Berghofer Medical Research Institute have collated a wide range of literature on a range of evolutionary adaptations. The review looks at how “Retooling the Protein Toolbox, Innovations in Regulatory RNA, Alterations to the Basic Genetic Code” and “Epigenetics: Fuelling Brain Plasticity and Adaptive Change” explains how our uniquely evolved human brain has come to be. Interestingly for this epigenetics focused audience, the authors site that an estimated 42% of human-chimpanzee gene expression differences are accounted for by epigenetic differences. The review combines “newly discovered genetic and epigenetic mechanisms with more established concepts” to create a more comprehensive picture of this fascinating field. For full text by Bitar and Barry see – https://academic.oup.com/mbe/article/35/2/263/4644722?searchresult=1
In a review that discusses the contribution of several cell death pathways to the life and death of activated T cells Zhan et al., highlight a mechanism of epigenetic regulation of cell survival unique to activated T cells. The review by Professor Lew and colleagues from the Walter and Eliza Hall Institute of Medical Research and University of Melbourne the collates the efforts made by studies aimed at understanding the survival and death of activated T cells.
For full text by Zhanet al., see – https://www.frontiersin.org/articles/10.3389/fimmu.2017.01809/full